Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase
- PMID: 26792518
- PMCID: PMC4763725
- DOI: 10.1073/pnas.1525055113
Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase
Abstract
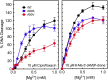
Mycobacterium tuberculosis is a significant source of global morbidity and mortality. Moxifloxacin and other fluoroquinolones are important therapeutic agents for the treatment of tuberculosis, particularly multidrug-resistant infections. To guide the development of new quinolone-based agents, it is critical to understand the basis of drug action against M. tuberculosis gyrase and how mutations in the enzyme cause resistance. Therefore, we characterized interactions of fluoroquinolones and related drugs with WT gyrase and enzymes carrying mutations at GyrA(A90) and GyrA(D94). M. tuberculosis gyrase lacks a conserved serine that anchors a water-metal ion bridge that is critical for quinolone interactions with other bacterial type II topoisomerases. Despite the fact that the serine is replaced by an alanine (i.e., GyrA(A90)) in M. tuberculosis gyrase, the bridge still forms and plays a functional role in mediating quinolone-gyrase interactions. Clinically relevant mutations at GyrA(A90) and GyrA(D94) cause quinolone resistance by disrupting the bridge-enzyme interaction, thereby decreasing drug affinity. Fluoroquinolone activity against WT and resistant enzymes is enhanced by the introduction of specific groups at the C7 and C8 positions. By dissecting fluoroquinolone-enzyme interactions, we determined that an 8-methyl-moxifloxacin derivative induces high levels of stable cleavage complexes with WT gyrase and two common resistant enzymes, GyrA(A90V) and GyrA(D94G). 8-Methyl-moxifloxacin was more potent than moxifloxacin against WT M. tuberculosis gyrase and displayed higher activity against the mutant enzymes than moxifloxacin did against WT gyrase. This chemical biology approach to defining drug-enzyme interactions has the potential to identify novel drugs with improved activity against tuberculosis.
Keywords: Mycobacterium tuberculosis; antibiotic resistance; complex stability; fluoroquinolones; gyrase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Similar articles
-
Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes.Antimicrob Agents Chemother. 2006 Jan;50(1):104-12. doi: 10.1128/AAC.50.1.104-112.2006. Antimicrob Agents Chemother. 2006. PMID: 16377674 Free PMC article.
-
Sensitivities of ciprofloxacin-resistant Mycobacterium tuberculosis clinical isolates to fluoroquinolones: role of mutant DNA gyrase subunits in drug resistance.Int J Antimicrob Agents. 2012 May;39(5):435-9. doi: 10.1016/j.ijantimicag.2012.01.007. Epub 2012 Mar 13. Int J Antimicrob Agents. 2012. PMID: 22421328
-
Role of gyrB Mutations in Pre-extensively and Extensively Drug-Resistant Tuberculosis in Thai Clinical Isolates.Antimicrob Agents Chemother. 2016 Aug 22;60(9):5189-97. doi: 10.1128/AAC.00539-16. Print 2016 Sep. Antimicrob Agents Chemother. 2016. PMID: 27297489 Free PMC article.
-
Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review.PLoS One. 2015 Mar 27;10(3):e0120470. doi: 10.1371/journal.pone.0120470. eCollection 2015. PLoS One. 2015. PMID: 25816236 Free PMC article. Review.
-
A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system.J Antimicrob Chemother. 2012 Apr;67(4):819-31. doi: 10.1093/jac/dkr566. Epub 2012 Jan 25. J Antimicrob Chemother. 2012. PMID: 22279180 Free PMC article. Review.
Cited by 33 articles
-
pH-driven enhancement of anti-tubercular drug loading on iron oxide nanoparticles for drug delivery in macrophages.Beilstein J Nanotechnol. 2021 Oct 7;12:1127-1139. doi: 10.3762/bjnano.12.84. eCollection 2021. Beilstein J Nanotechnol. 2021. PMID: 34703723 Free PMC article.
-
Screening of Compounds for Anti-tuberculosis Activity, and in vitro and in vivo Evaluation of Potential Candidates.Front Microbiol. 2021 Jun 30;12:658637. doi: 10.3389/fmicb.2021.658637. eCollection 2021. Front Microbiol. 2021. PMID: 34276592 Free PMC article.
-
Elucidating the Antimycobacterial Mechanism of Action of Ciprofloxacin Using Metabolomics.Microorganisms. 2021 May 28;9(6):1158. doi: 10.3390/microorganisms9061158. Microorganisms. 2021. PMID: 34071153 Free PMC article.
-
Piperidine-4-Carboxamides Target DNA Gyrase in Mycobacterium abscessus.Antimicrob Agents Chemother. 2021 Jul 16;65(8):e0067621. doi: 10.1128/AAC.00676-21. Epub 2021 Jul 16. Antimicrob Agents Chemother. 2021. PMID: 34001512 Free PMC article.
-
Recent Progress and Challenges for Drug-Resistant Tuberculosis Treatment.Pharmaceutics. 2021 Apr 21;13(5):592. doi: 10.3390/pharmaceutics13050592. Pharmaceutics. 2021. PMID: 33919204 Free PMC article. Review.
Publication types
MeSH terms
Substances
Grant support
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

