Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements
Abstract
Frontotemporal dementia with parkinsonism, chromosome 17 type (FTDP-17) is caused by mutations in the tau gene, and the signature lesions of FTDP-17 are filamentous tau inclusions. Tau mutations may be pathogenic either by altering protein function or gene regulation. Here we show that missense, silent, and intronic tau mutations can increase or decrease splicing of tau exon 10 (E10) by acting on 3 different cis-acting regulatory elements. These elements include an exon splicing enhancer that can either be strengthened (mutation N279K) or destroyed (mutation Δ280K), resulting in either constitutive E10 inclusion or the exclusion of E10 from tau transcripts. E10 contains a second regulatory element that is an exon splicing silencer, the function of which is abolished by a silent FTDP-17 mutation (L284L), resulting in excess E10 inclusion. A third element inhibiting E10 splicing is contained in the intronic sequences directly flanking the 5′ splice site of E10 and intronic FTDP-17 mutations in this element enhance E10 inclusion. Thus, tau mutations cause FTDP-17 by multiple pathological mechanisms, which may explain the phenotypic heterogeneity observed in FTDP-17, as exemplified by an unusual family described here with tau pathology as well as amyloid and neuritic plaques.
Frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) is an autosomal-dominant disease with variable clinical and neuropathologic features (1). Symptoms can include personality changes sometimes with psychosis, hyperorality, reduced speech output, and loss of executive function (2–8). Memory is retained until late in the disease (9). Parkinsonism and amyotrophy occur in some families (2). Neuropathologic changes include frontotemporal atrophy, sometimes with atrophy of the basal ganglion, substantia nigra, and amygdala. FTDP-17 is caused by mutations in the gene for tau (10–12). Tau is a microtubule-associated protein that normally functions to promote microtubule (MT) assembly and stability. In FTDP-17, tau aggregates in the brain to form abnormal filamentous structures including neurofibrillary tangles (NFTs) (9, 13, 14), neuropil threads, glial tangles, and dense intracellular deposits (5). The type and location of tau pathology varies between different FTDP-17 families.
Tau mutations cause FTDP-17 by at least two different mechanisms. First, intronic mutations immediately adjacent to the 3′ end of alternatively spliced exon 10 (E10), increase inclusion of this exon in tau transcripts (10). E10 encodes one of four nearly identical MT-binding motifs found in the longer isoforms of tau. When E10 is included, isoforms with four MT binding domains (four-repeat or 4R tau) are produced, and when E10 is excluded, tau isoforms with three MT repeats (three-repeat or 3R tau) are produced. Tau FTDP-17 mutations that increase E10 result in the production of excess 4R tau as observed in both soluble and insoluble tau from FTDP-17 autopsy samples (15).
A second mechanism by which tau mutations cause FTDP-17 is by impairing tau protein function. Tau with G272V, P301L, V337M, or R406W mutations exhibits reduced affinity and capacity for MT-binding and a reduced ability to facilitate MT polymerization when compared with normal tau (15, 16). However, not all missense mutations alter the ability of tau to interact with MTs (e.g., mutation N279K)(15, 17). The present study was undertaken to determine how FTDP-17 mutations affect gene regulation and tau protein function and to identify the mechanisms by which some of these mutations alter regulation of E10 splicing.
METHODS
Subjects.
The LKL family proband (III-4) is a 53-year-old machinist with disease onset at age 49. Initial symptoms were forgetting recent conversations and becoming lost while driving. At age 51, his Mini Mental Status Exam (MMSE) score was 28/30. He had only mild word-finding difficulties. His general neurological examination was normal. An MRI brain scan showed mild anterior temporal atrophy. At age 53 his MMSE was 25/30. Detailed neuropsychometric testing revealed intact simple and divided attentional abilities with impaired language, visuospatial, and visuoconstructional abilities. Executive functions were in the borderline-to-impaired range.
There have been six affected persons in this family. Mean age-of-onset is 51.8 ± 4.8 years. Typical symptoms have been forgetfulness, confusion, lack of insight, and poor judgement and executive function. Three persons have required psychiatric hospitalization.
The mother of III-6 is an 88-year-old woman (II-2S) who is currently institutionalized with a diagnosis of late-onset Alzheimer’s disease (AD) (onset approximately age 75). A computerized tomography brain scan has shown mild generalized cerebral atrophy and her apolipoprotein E genotype is 4/4.
Mutation Screening.
Genomic DNA was amplified by using primers complementary to intronic sequences flanking each exon for tau (12), presenilin 1 (18), and presenilin 2 (19). For the amyloid precursor protein gene, exons 16 and 17 were screened (20) because all amyloid precursor protein mutations identified to date are in these two exons. Both strands of each exon were sequenced. Amplified DNA was purified by using gel electrophoresis and Gene-Clean (Bio 101). Fragments were sequenced by using TaqFS DNA polymerase, flourescent dye terminators, and an Applied Biosystem ABI377 DNA sequencer.
Splicing Assays.
An E10 fragment containing 33 bp and 51 bp of flanking intron sequences was generated by PCR amplification of PAC 4139 using the PCR oligonucleotide primers I9X2 and I10B2 (5′-CCACTCGAGCGTGTCACTCATCCTTTTTTC-3′ and 5′-CGGGATCCCCTAATAATTCAAGCCACAG-3′, respectively). The PCR product was digested with XhoI and BamHI and inserted into vector pSPL3 (21) (GIBCO/BRL). Mutations were generated by PCR using a mutagenized oligonucleotide together with primer I9X2 and I10B2. All constructs were sequenced before transfection. COS-7 cells were maintained in 10% fetal bovine serum and DMEM, and transfected by using Lipofectamine (GIBCO/BRL) according to manufacturer’s instructions. Total cellular RNA was isolated 48 hours posttransfection by using TRIzol (GIBCO/BRL). RNA samples were DNase 1 (Amersham Pharmacia) treated before reverse transcription. RNA (2.5 μg) was reverse-transcribed by using the GeneAmp RNA PCR kit (Perkin–Elmer). Splicing products were PCR-amplified by using pSPL3 vector-specific primers SD6 (5′-TCTGAGTCACCTGGACAACC-3′) and SA2 (5′-ATCTCAGTGGTATTTGTGAGC-3′, 32P-labeled). PCR was performed for 18 cycles to obtain linear amplification. Products were resolved on 5% acrylamide gels and quantitated by using a phosphoimager.
In Vitro Expression and Purification of Wild-Type and Mutant Tau Proteins.
E10 missense mutations were introduced by site-directed mutagenesis into PRK172/T40, which contains the longest human brain tau isoform with exons 2,3, and 10 but not 4a, 6, or 8. Recombinant proteins for functional studies were produced in Esherichia coli and purified by mono-S FPLC. The protein concentrations were determined by protein assay using BCA reagents (Pierce), densitometry of Coomassie-stained gels, and quantitative Western blotting by using polyclonal antiserum to tau (17026) detected with 125I-labeled protein A.
MT-Binding Assay.
MTs were assembled from phosphocellulose-purified bovine tubulin (Cytoskeleton, Denver) in reassembly (RA) buffer (0.1 M MES, pH 6.8/0.5 mM MgSO4/1 mM EGTA/2 mM DTT/mixture of protease inhibitors) containing 1 mM GTP and 20 μM Taxol as described (22). Three micromolar Taxol-stabilized MT dimers were incubated with 0–1.5 μM recombinant tau at 37°C for 20 minutes. A layer of sucrose cushion was underlaid, and the bound tau and free tau were separated by centrifugation at 50,000 × g (25°C) for 20 minutes. Bound and free tau were determined by quantitative Western blotting using polyclonal antiserum to tau (17026) detected with 125I-labeled protein A. Each experiment was repeated at least three times with at least two different recombinant tau preparations.
Light-Scattering Assay to Assess Tau-Promoted MT Assembly.
Bovine tubulin monomers (35 μM) was mixed at 4°C with 15 μM recombinant tau in RA buffer supplemented with 1 mM GTP (23). Samples were gradually warmed to 37°C at a controlled rate in quartz cuvettes, and turbidity was monitored by reading the optical density at 350 nm (A350) with a Beckman DU640 spectrophotometer at 1-minute intervals. Duplicate samples were examined in each experiment, which was repeated at least three times with at least two different recombinant tau preparations.
RESULTS
Tau Mutations and FTDP-17 Neuropathology.
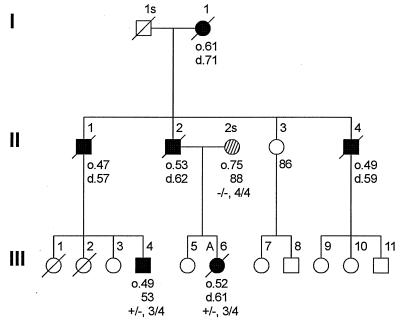
The mutations tested are those clustered in or near E10 (Table (Table1;1; Fig. Fig.1)1) and include a silent mutation (L284L) identified in the LKL family (Fig. (Fig.2).2). This mutation cosegregates with the disease in this family and was not present in 96 controls. No other mutations were found in LKL subjects in other tau exons, in presenilin 1 or 2, or in the amyloid precursor protein gene exons that encode β-amyloid (Aβ) protein.
Table 1
Tau mutations and polymorphism
| Mutation | Location | Domain in tau protein or gene | Nucleotide change | Ref. |
|---|---|---|---|---|
| N279K | E10 | IR1-2 | AAT → AAG | 17 |
| Δ280K | E10 | IR1-2 | 3-bp deletion | 24 |
| L284L | E10 | IR1-2 | CTT → CTC | This study |
| P301L | E10 | R2 | CCG → CTG | 10, 12, 17 |
| S305N | E10/5′ splice site | IR2-3 | AGT → AAT | 25 |
| E10 + 3 | I10 | 5′ splice site | g → a | 11 |
| E10 + 14 | I10 | Intronic | c → t | 10 |
| E10 + 16 | I10 | Intronic | c → t | 10 |
| E10 + 29 | I10 | Intronic | g or a | This study |
Inter repeat (IR) regions are sequences between the MT-binding domains (e.g., IR1–2 is between MT binding domains 1 and 2). The sites of I10 mutations are designated by the number of the intronic base that is mutated. E10 + 29 is a polymorphism with either an G or A at intronic nucleotide 29 with allele frequencies of 0.98 and 0.02, respectively, as determined in a Caucasian population (n = 96).
E10/I10 tau mutations. RNA sequences for part of E10 (A) and the E10/I10 junction (B) are shown. Capital letters are E10, and lower case letters are I10. The hatched box below the E10 sequence spans the proposed purine-rich exon splicing enhancer. The hatched box above the sequence spans the proposed exon splicing silencing sequence. The normal tau E10/I10 junction sequence is presented as a potential stem–loop. The E10 + 16 mutation was used in splicing experiments with the 305N mutation to attempt to restore putative stem–loop base pairing. Likewise, an E10 + 12 substitution was used in double-mutant constructs to compensate for the E10 + 3 mutation. The predicted free energies in kcal/mol (1 kcal = 4.18 J) at 37°C are: −9.2, normal (stem–loop shown); −6.8, 305N; −7.8, 305N//E10 + 16; −4.5, E10 + 3; −7.1, E10 + 3/E10 + 12; −6.7, E10 + 14; −7.8, E10 + 16 using the GCG version of mfold (39).
The LKL pedigree. Circles are females, squares are males, and deceased subjects have a line through the subject symbol. Filled symbols are subjects with frontotemporal dementia, and the hatched circle is a probable AD subject. Current age is below affected subjects, with age-of-onset indicated with an “o” and age-at-death with a “d.” Subjects heterozygous for the L284L mutation are shown as “+/−,” and subjects homozygous for the normal allele are shown as “−/−.” Apolipoprotein E genotypes are shown as 3/4 or 4/4 (no other apolipoprotein E genotypes were observed). “A” indicates an autopsy was performed.
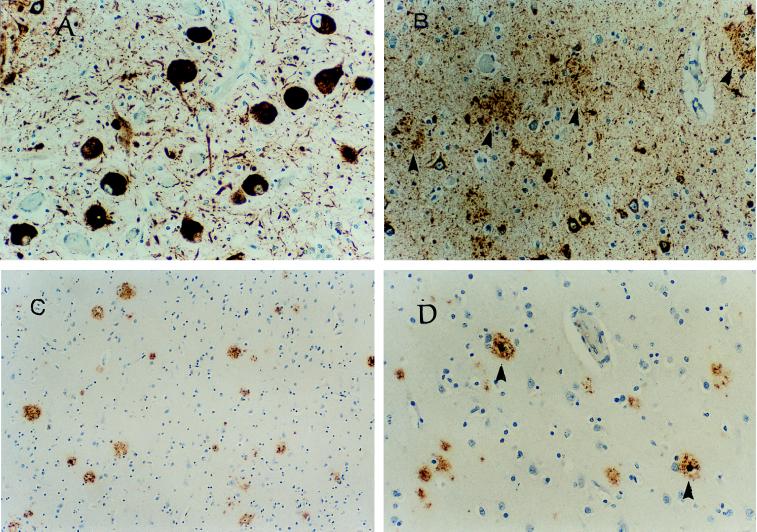
Affected subjects from the LKL family have a variant of frontotemporal dementia with difficulties in word-finding and visual-spatial abilities, behavioral changes, and abnormal executive function. An autopsy from a family member (III-6, Fig. Fig.2),2), who had disease onset at age 52 and died at age 61 showed both tau pathology and amyloid deposits. The brain weighed 941 grams with moderate bilateral frontal and right temporal atrophy and an old infarction in the right superior temporal–inferior parietal region. NFTs ranged from sparse to frequent in the neocortex, amygdala, and parahippocampus. The hippocampus showed sparse NFTs but large numbers of argyrophilic grains and neuropil threads from CA1 to subiculum and parahippocampus. Immunostaining for tau by using tau-2 antibody showed large numbers of tau-positive neurons with two types of deposits: cytoplasmic granular material and dark, dense bundles consistent with flame-shaped or globose NFTs. Both types of tau aggregates were in the neocortex, amygdala, hippocampus, parahippocampus, basal ganglia, substantia innominata, substantia nigra, red nucleus, and brainstem raphe (Fig. (Fig.33A). Numerous neuropil threads and grains were in the gray matter. Glial cells were tau-positive in the white matter but not in the optic tract. In the cortex, tau-2 staining showed a laminar distribution with relative sparing of layer III. Tau-positive neurons were present in the parahippocampus and to a lesser extent in the hippocampus. The substantia nigra had a large number of tau-positive neurons. There were clusters of tau-positive neurites scattered about the neocortex and limbic areas consistent with neuritic plaques (Fig. (Fig.33B). Aβ immunostaining showed numerous diffuse plaques in the neocortex as well as a smaller number of neuritic plaques with dense cores (Fig. (Fig.33 C and D). The regional plaque distribution was not significantly different from typical cases of late-onset AD. Overall, the amyloid content and other neuropathological findings met the Khachaturian (26), Consortium to Establish a Registry for Alzheimer’s Disease (CERAD), (27) and National Institute of Aging/Reagan (28) criteria for AD, although the most profound pathology was tau deposits in neurons and neuropil threads.
Autopsy results from LKL subject III-6. (A) Tau-2 immunopositive neurons in the substantia inomminata. Dense, tau-positive, cytoplasmic inclusions are seen in multiple neurons as well as neuropil threads (magnification ×219). The section was stained by antibody tau-2 (Sigma, diluted 1:12,000) with ABC-peroxidase-diaminobenzidine and counterstained with hematoxylin. (B) Dystrophic processes of neuritic plaques, neurofibrillary tangles, and neuropil threads in the entorhinal cortex. Staining was with tau-2 as in A (magnification ×219). (C) Aβ-positive plaques with and without dense cores in superior temporal cortex (magnification ×109). Staining was with anti-Aβ antibody 10D5 (Athena Neurosciences, San Francisco, diluted 1:5,000) as described in A above. (D) Higher magnification of superior temporal cortex showing two Aβ-positive plaques with dense cores (magnification ×219). Staining was as in C above.
Tau Gene Regulation.
The effects of tau mutations on E10 splicing were assayed by inserting normal or mutant tau E10 with 33 bp of flanking intron 9 (I9) and 51 bp of flanking intron 10 (I10) into the intron of a minigene constructed from heterologous intron and exon sequences (21). When normal E10 is tested, both E10+ and E10− transcripts are produced, with E10+ representing 37.7% of the total (Fig. (Fig.4).4). For mutations in the E10 coding sequence, two missense mutations, 279K and 305N, increased inclusion of E10 to 79.1% and 80.6%, respectively. The silent mutation 284L also produces an increase in E10+ transcripts resulting in almost no E10− transcript. In contrast, the Δ280K deletion results in no detectable E10+ transcripts. The 301L mutation did not alter E10 splicing.
Affects of E10 mutations on splicing. (A) Autoradiograph of reverse transciption–PCR products from splicing assays. E10− and E10+ transcripts yield 261-bp and 354-bp fragments, respectively. For each construct, where sufficient quantities of each band were present, both E10+ and E10− fragments were excised from gels, and the DNA sequence was determined. In each case, the splicing occurred at the expected sites. (B) Quantitation of E10− and E10+ splicing. Each bar represents the mean of five separate transfection experiments, and 100% is the sum of E10− and E10+. Error bars are the standard deviations. For the comparison of normal tau to mutations, ∗ indicates P < 0.000001. Mutation 301L and polymorphism E10 + 29A were not significantly different from normal tau.
Mutations in the first 16 bp of I10 also induce oversplicing (10). We confirmed this observation and extended it to show that mutation E10 + 3 also causes oversplicing (Fig. (Fig.4).4). Based on the consensus sequence for 5′ splice sites, this mutation does not obviously strengthen the E10 5′ splice site in terms of U1 small nuclear ribonucleoprotein or U6 small nuclear ribonucleoprotein binding (29, 30). Thus the E10 + 3 mutation must affect splicing by some other mechanism. One proposal (10, 11) is that a stem–loop structure forms at the E10/I10 junction that inhibits E10 inclusion by interfering with U1snRNP hybridization. I10 mutations would destabilize this structure by reducing by 1 the number of potential base pairs in the stem (Fig. (Fig.1).1). To test this hypothesis, compensatory double mutants were generated so that the base pairing in the stem–loop destroyed by FTDP-17 mutations are restored by replacing a normal nucleotide with one that is complementary to the mutant base (e.g., E10 + 3 plus E10 + 12 c → t change, Fig. Fig.1).1). The E10 + 3 mutation was selected because the normal and mutant sequence both match the 5′splice site consensus sequence for constitutively spliced exons. The double mutant E10 + 3/E10 + 12c overspliced E10 to similar levels as E10 + 3 alone, and normal splicing was not restored (Fig. (Fig.4).4). Similar experiments were not performed for E10 + 14 or E10 + 13 because the compensatory changes at E10 + 1 and E10 + 2, respectively, would destroy the 5′ splice site by altering invariant nucleotides required for splicing (31). A double mutant with a compensatory change opposite 305N also overspliced. In fact, the 305N/E10 + 16 double mutant actually produces more E10+ transcripts than the 305N mutation alone, indicating that the E10/I10 junction inhibitory element still functions when the 5′ splice site is strengthened by the 305N change. Therefore, it seems that disruption of the putative stem–loop is not the mechanism through which these intronic mutations alter E10 splicing. During screening for mutations, a polymorphism in I10 was identified (E10 + 29). This polymorphism did not affect splicing.
Tau Protein Function.
The effects of E10 mutations on tau protein function was tested in MT-binding and MT-polymerization assays (Fig. (Fig.5).5). For 301L and Δ280K, the maximal amount of tau bound (Bmax) to MTs was decreased and the affinity of tau for MTs was reduced (Table (Table2).2). In contrast, MT binding by mutant 279K tau and 305N tau was the same as for normal tau. MT assembly was measured under conditions in which no polymerization occurs unless tau is present (Fig. (Fig.55B). This assay measures the lag time required to nucleate MTs, the rate of tau-induced MT assembly, and the maximal amount of MT polymers formed (Table (Table2).2). As in the MT binding assay, 279K and 305N were indistinguishable from normal tau. However, the Δ280K and 301L mutant tau proteins nucleate MTs less efficiently (compare lag times of 5 min for Δ280K and 301L with 2 min for normal tau, Table Table2).2). The maximal MT polymer mass formed in the presence of Δ280K was also significantly lower than that of normal tau (Table (Table2).2). Thus, 301L and Δ280K appear to alter tau protein function, whereas 279K and 305N do not. The latter two mutants appear to act entirely by affecting E10 splicing (Fig. (Fig.4).4).
The Δ280K and 301L, but not 279K or 305N, mutations impair the functions of tau in vitro. (A) Tau binding to MTs. Binding curves were generated by fitting data using nonlinear regression with the standard binding equation: (bound tau) = Bmax × (free tau)/[Kd+(free tau)]. The Bmax and Kd values are listed in Table Table2.2. Symbols: ■, normal; ▵, 279K; ∗, Δ280K; ⋄, 301L; ●, 305N. (B) Tau-promoted MT assembly. Bovine tubulin monomers (35 μM) were mixed at 4°C with 15 μM recombinant tau in RA buffer supplemented with 1 mM GTP. Samples were warmed to 37°C, and MT assembly was monitored by optical density at 350 nm (A350). A representative result is shown and the lag times, initiation rates, and A350 max values (n = 6) are summarized in Table Table2.2. Symbols are as in A; ♦ indicates no added tau.
Table 2
MT-binding- and MT assembly-promoting properties of normal and mutant tau
| MT binding
| MT assembly promotion
| ||||
|---|---|---|---|---|---|
| Bmax, μM* | Kd, μM* | Lag time, ∼min | Initiation rate, OD/min | A350 max | |
| Normal | 1.238 ± 0.042 | 0.039 ± 0.006 | 2 | 0.15 | 0.534 ± 0.014 |
| 279K | 1.265 ± 0.055 | 0.056 ± 0.012 | 2 | 0.16 | 0.557 ± 0.005 |
| Δ280K | 1.011 ± 0.080 | 0.1526 ± 0.035† | 5 | 0.14 | 0.472 ± 0.006† |
| 301L | 0.883 ± 0.047† | 0.079 ± 0.016‡ | 5 | 0.14 | 0.518 ± 0.012 |
| 305N | 1.184 ± 0.166 | 0.063 ± 0.026 | 2 | 0.15 | 0.529 ± 0.011 |
Bmax, kd, values are given as mean ± SEM.
DISCUSSION
Our work shows that FTDP-17 tau mutations can either increase or decrease E10 splicing and that regulation of E10 splicing is complex involving at least 3 different cis-acting elements. Mutation 279K increases E10 inclusion presumably by enhancing an existing or creating a new exon splicing enhancer element. The fact that the in-frame deletion of the 3 purines in the adjacent codon (Δ280K) abolishes E10 splicing strongly suggests that 279K and Δ280K both affect an existing exon splicing enhancer. The 279K change may enhance splicing by increasing the purine content of this exon splicing enhancer, or by creating a GAR repeat (where R is a purine) which, when present in two or more copies in other genes, enhances splicing (32–35). The 279K-induced increase in splicing is consistent with previous work showing that the 4R/3R tau protein ratio is increased in brains from subjects with this mutation (15, 17). The silent mutation 284L increases E10+ transcripts, presumably by destroying an exon splicing silencing element. The normal tau sequence, UUAG, affected by this mutation is an exon splicing silencing in the HIV-I tat exon 3, which suppresses exon inclusion (36). Mutation 305N is within the 5′ splice site recognition sequence for E10 (Fig. (Fig.1)1) and presumably causes oversplicing by changing the normally weak tau E10 5′ splice site sequence of GUgugagu (capital letters are E10, lower case letters are intron 10 or I10) to AUgugagu, which is a stronger splice site and a better match of the 5′ consensus sequence of AGgu(a/g)agu for a constitutively included exon (31).
The I10 mutations presumably act to disrupt an inhibitory element immediately adjacent to E10. However, our results do not support the role of a short stem–loop in this process (10, 11). The E10 + 3 mutation is particularly intriguing because it should not alter the strength of the 5′ splice site because an A or G is equivalent at this site (31). Inhibition of splicing by I10 sequences may be caused by some other higher order structure that does not involve the proposed stem–loop or may be the result of specific regulatory factors that bind to this region and sterically hinder recognition or usage of the 3′ end of the exon by splicing factors.
The data presented here and in previous studies (15, 16) show that most missense mutations alter the interactions of tau with MTs. These mutations may cause FTDP-17 either by impairing normal MT function, or the reduced MT binding may result in an increase in 4R tau available for aggregation. The Δ280K mutation is enigmatic in that it affects both gene regulation and the biochemical properties of the tau protein. The functional consequence could be that although 4R tau levels are reduced, sufficient Δ280K tau is present to trigger disease. Alternatively, 4R and 3R MT binding may be tightly coupled, and an excess of either tau isoform could result in excess free tau and subsequent aggregation. This would be analogous to Pick’s disease, where abnormal tau aggregates are predominantly 3R tau (37).
The extensive phenotypic heterogeneity seen in FTDP-17 is presumably the consequence of different mutations acting by different mechanisms. The V337M mutation (12) results in NFTs and paired helical filaments that are indistinguishable from AD, and no tau glial pathology is present (9, 13). In contrast, for the E10 + 3 mutation, both neuronal and glial tau aggregates are present, and the filaments formed are predominantly straight (5). The LKL family presented here, like the E10 + 3 kindred, showed a variety of tau aggregates in both neurons and glial cells (Fig. (Fig.3).3). However, unlike other FTDP-17 subjects from other families, a substantial number of diffuse and neuritic Aβ plaques were found. These results must be interpreted with caution because only a single autopsy is available and coincident AD and FTDP-17 cannot be ruled out. Nonetheless, this subject died at a relatively young age (61 years), when AD is rare and plaques are infrequent in controls. These results raise the intriguing possibility that under certain circumstances, mutation-induced abnormal tau can result in Aβ deposition, which is consistent with the hypothesis that the first observable pathology in AD may be NFTs in the entorhinal cortex (38). The L284L mutation causing this unique pathology is the only known mutation affecting the E10 exon splicing silencing element, and this element probably binds to an unidentified trans-acting inhibitory factor(s). Thus, the differential expression of this factor(s) may determine the unique pathology observed here.
Acknowledgments
We thank Dr. John Trojanowski for critical reading of the manuscript and Elaine Loomis, LeoJean Anderson, Randy Small, and Christiane Ulness for technical support. This work was funded by National Institute of Aging Grant AG11762 (G.D.S.), Veterans Affairs Merit Review awards (T.D.B. and G.D.S.), and AG10210 (V.M.-Y.L.) and a National Institute of Aging-funded Alzheimer’s Disease Research Center (AG05136). Informed consent was obtained from each subject or next-of-kin with approval of the University of Washington Human Subjects Review Committee.
ABBREVIATIONS
| 3R | 3-repeat tau |
| 4R | 4-repeat tau |
| AD | Alzheimer’s disease |
| E10 | tau exon 10 |
| FTDP-17 | frontotemporal dementia with parkinsonism-chromosome 17 type |
| MT | microtubule |
| NFT | neurofibrillary tangle |
| Aβ | β-amyloid protein |